Google AI unleashes Deeppolisher: a new deep learning tool that improves the accuracy of genomic components by precisely correcting basic-level errors
Google AI collaborates with UC Santa Cruz Genome Academy to introduce DeeppoliserThis is a cutting-edge deep learning tool designed to essentially improve the accuracy of genomic components by correcting basic levels of errors. Its significant efficacy has been demonstrated recently in promoting human pangenome reference, a major milestone in genomic research.
The challenge of accurate genome assembly
The reference genome is the basis for understanding genetic diversity, heredity, disease mechanisms, and evolutionary biology. Modern sequencing technologies, including those developed by Illumina and Pacific Biosciences, have greatly improved sequencing accuracy and throughput (even with technological breakthroughs, assembling error-free human genomes (more than 3 billion nucleotides) remains very challenging. Even tiny error rates per bond can lead to thousands of errors that can mask critical genetic mutations or mislead downstream analyses.
What is Deeppolisher?
Deeppoliser It’s one Open source, transformer-based sequencing correction tool. Building on the advances of DeepConsensus, it utilizes Transformer Deep Leaver Architectures to further reduce genome assembly errors, especially Insert and Delete (Indel) ErrorsThis can have profound effects by transferring the reading framework and can lead to the missing of important genes or regulatory elements during annotation.
- technology: Encoding transformer only, adapts to reliable technology in genomics natural language processing.
- Training data: Human cell lines characterized by NIST and NHGRI were utilized and sequenced with various platforms to ensure near-complete accuracy (~99.999999% correctness, between 300-1,000 errors in 6 billion bases).
How does it work? (Technical Overview)
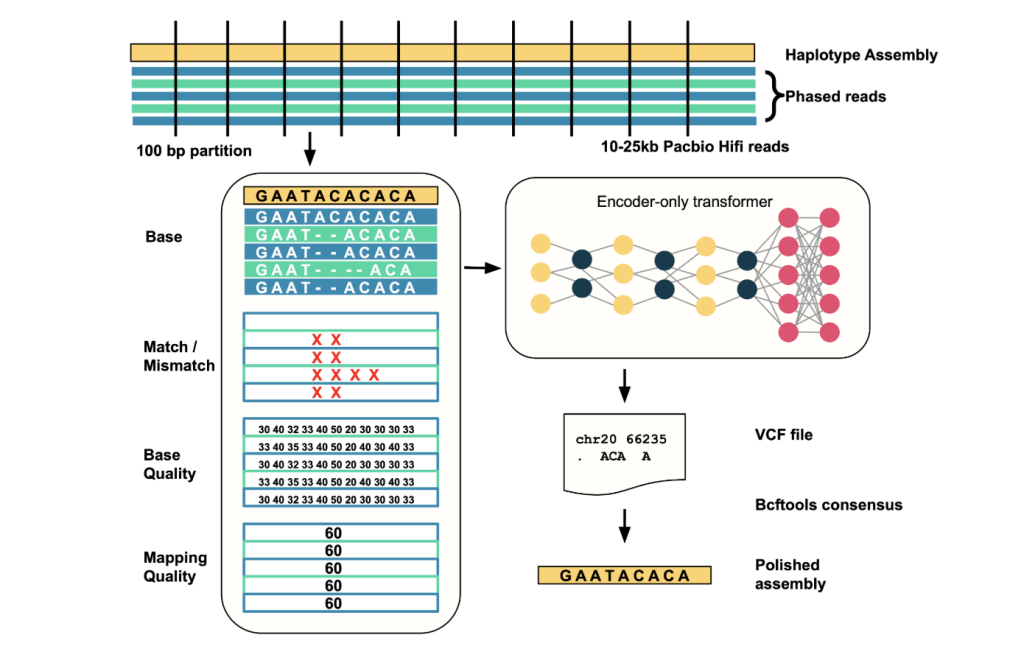
- Input Alignment: Aligned PACBIO HIFI read haplotype-resolved genomic components as input.
- Error site detection: Scan the component in a 25KB window; identify candidate error sites where reading evidence deviates from the component.
- Data encoding: For each window containing presumed errors (
- Model inference: These tensors are fed into the transformer, which predicts the correction sequence of these regions.
- Output correction: Output the difference in VCF format and then apply it to the component to generate a polished highly accurate sequence using a tool like BCFTool.

Performance and impact
Deeppolisher provides substantial improvements:
- Reduce total error: ~50%
- Reduce the Indel Error: > 70%
- Error rate: Through the Human Pangenome Reference Alliance (HPRC), the error rate for the basic basis for every 500,000 assembled bases is as low as one basic error in actual deployment.
- Improvements to genomic Q score: The assembly quality is increased from Q66.7 to Q70.1 on average (Q score is a logarithmic measure of the error rate per bit; higher rates are better. Q70.1 means
- Each sample is tested Improvement was shown by HPRC.
These advances directly affect the reliability and accuracy of derived citations, e.g. People pangenome referenceit sees five times the data expansion and a large amount of error reduction due to fur caterpillars.


Deployment and Applications
- Integrate in the main project: In the second data release for HPRC, highly accurate reference components were provided for 232 individuals, ensuring a wide range of ancestral diversity in genomic references.
- Open Source Access: Obtained through GitHub, case studies and docking workflows, used for components generated by tools such as Hifiasm, and sequenced with PACBIO HIFI reads.
- Generalization: Although initially focused on the human genome, structures and methods adapted to other biological and sequencing platforms, thus promoting accuracy across the genomic community.
Practical workflow examples
A typical workflow using Deeppolisher might involve:
- Input: Hifiasm diploid component and PACBIO HIFI read, phased using pharaoh pipe.
- Run: dockerized commands for image creation, reasoning and correction applications.
- Output: VCF files for parent and father components individually, polishing FASTAS after BCFTOOLS consensus step.
- Evaluation: Use benchmarking tools (e.g., DIPCALL, HAP.PY) to quantify the improvement of error rate and change accuracy.
Conclusion and future direction
Deeppoliser Represents a leap in genomic polishing technology – reducing error rates and unlocking higher resolutions for functional genomics, rare variant discovery and clinical applications. By targeting the remaining barriers to perfect genome combinations, it can enable more accurate diagnosis, population-level genetic research and pave the way for next-generation reference projects that benefit from biomedical research and medicine.
Check Technical details,,,,, Github page and Paper. Check out ours anytime Tutorials, codes and notebooks for github pages. Also, please stay tuned for us twitter And don’t forget to join us 100K+ ml reddit And subscribe Our newsletter.
Asif Razzaq is CEO of Marktechpost Media Inc. As a visionary entrepreneur and engineer, ASIF is committed to harnessing the potential of artificial intelligence to achieve social benefits. His recent effort is to launch Marktechpost, an artificial intelligence media platform that has an in-depth coverage of machine learning and deep learning news that can sound both technically, both through technical voices and be understood by a wide audience. The platform has over 2 million views per month, demonstrating its popularity among its audience.


