Google AI Research Releases DeepSomatic: A New AI Model for Identifying Genetic Variations in Cancer Cells
A team of researchers from Google Research and the University of California, Santa Cruz released deep somatic cellsan artificial intelligence model that identifies genetic variations in cancer cells. In the Children’s Mercy study, it found 10 variants in children’s leukemia cells that other tools missed. DeepSomatic has a somatic small variant calling program for cancer genomes that works on Illumina short reads, PacBio HiFi long reads, and Oxford Nanopore long reads. The method extends DeepVariant to detect single nucleotide variants and small insertions and deletions in whole-genome and whole-exome data, and supports tumor-normal and tumor-only workflows, including FFPE models.
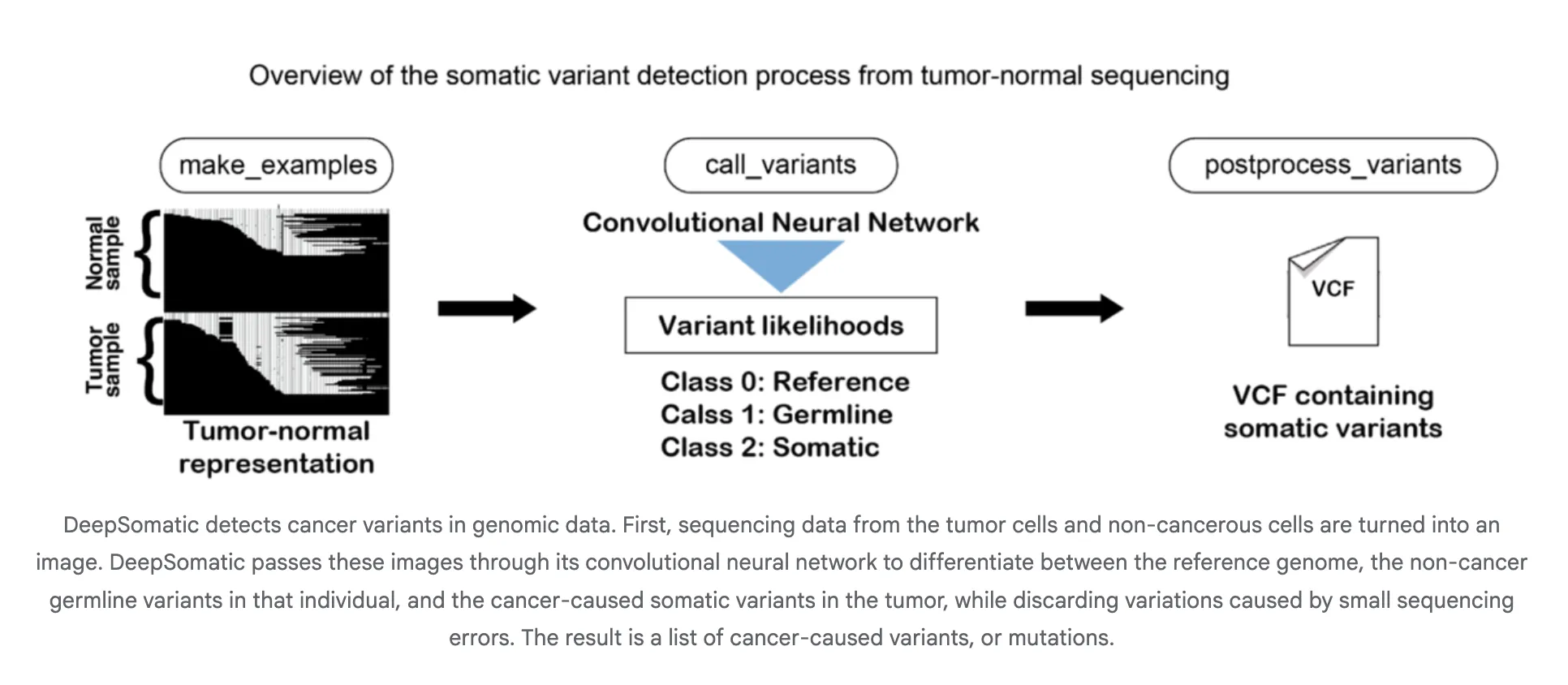
How does it work?
DeepSomatic converts aligned reads into images, such as tensors encoding stacking, elementary mass, and alignment context. A convolutional neural network classifies candidate sites as somatic or non-somatic, and the pipeline emits VCF or gVCF. This design is platform-agnostic as the tensor summarizes local haplotypes and error patterns across technologies. Google researchers describe the method and its focus on distinguishing inherited and acquired variants, including in difficult samples such as glioblastoma and childhood leukemia.
Datasets and benchmarks
Training and assessment using CASTLE, Cancer Standards Long Read Assessment. CASTLE contains 6 matched pairs of tumor and normal cell lines that were whole-genome sequenced on Illumina, PacBio HiFi, and Oxford Nanopore. The research team released the benchmark set and materials for reuse. This fills a gap in multi-technology somatic training and testing resources.
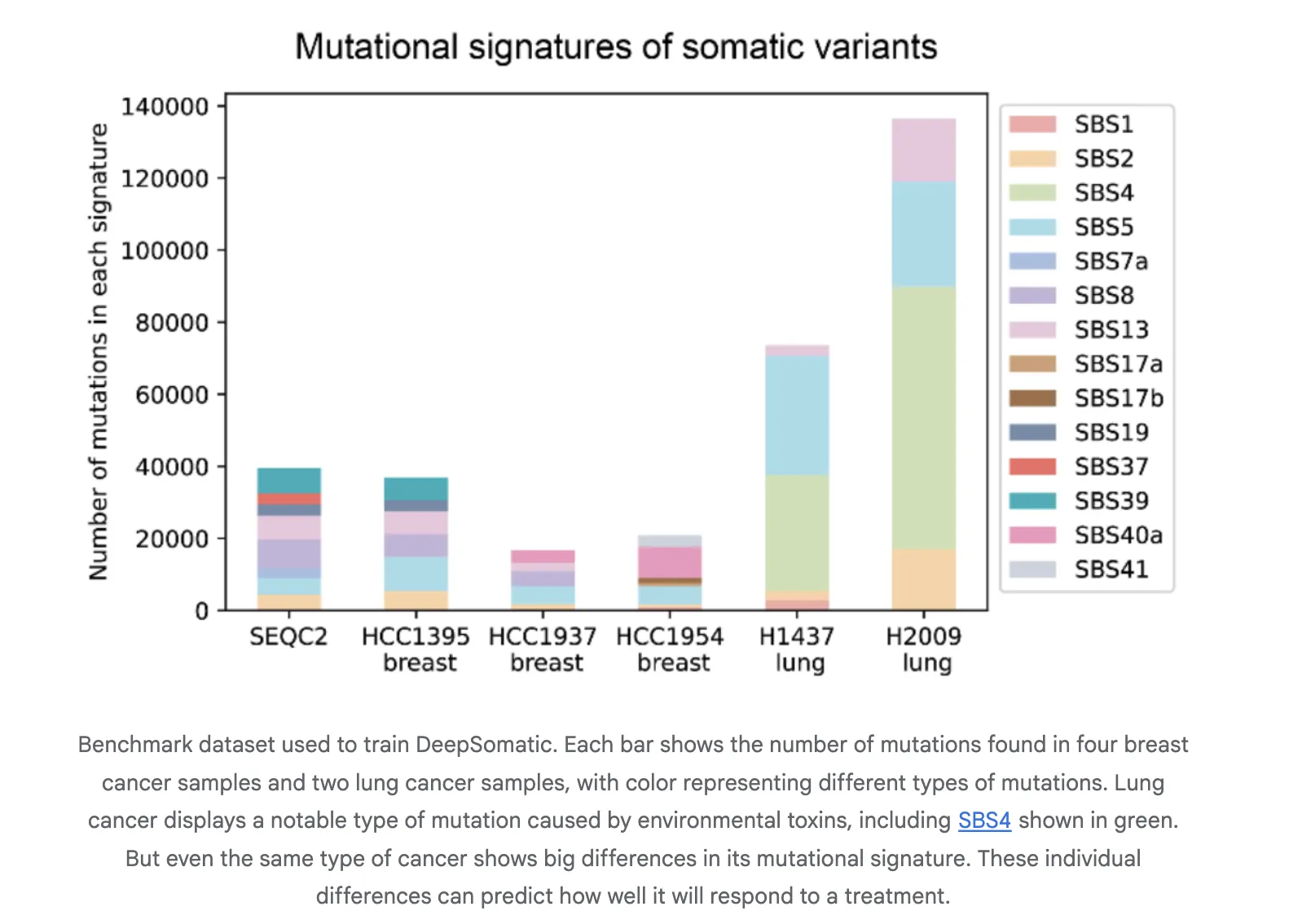

Report results
The team reported consistent results compared with widely used methods on single nucleotide variations and indels. On Illumina indels, the next best method is about 80% F1 and DeepSomatic is about 90%. On PacBio indels, the next best method is below 50% and DeepSomatic is above 80%. Baselines include SomaticSniper, MuTect2 and Strelka2 for short reads and ClairS for long reads. The study reported 329,011 somatic variants in the reference line as well as additional preserved samples. The Google research team reports that DeepSomatic outperforms current methods, especially for indels.
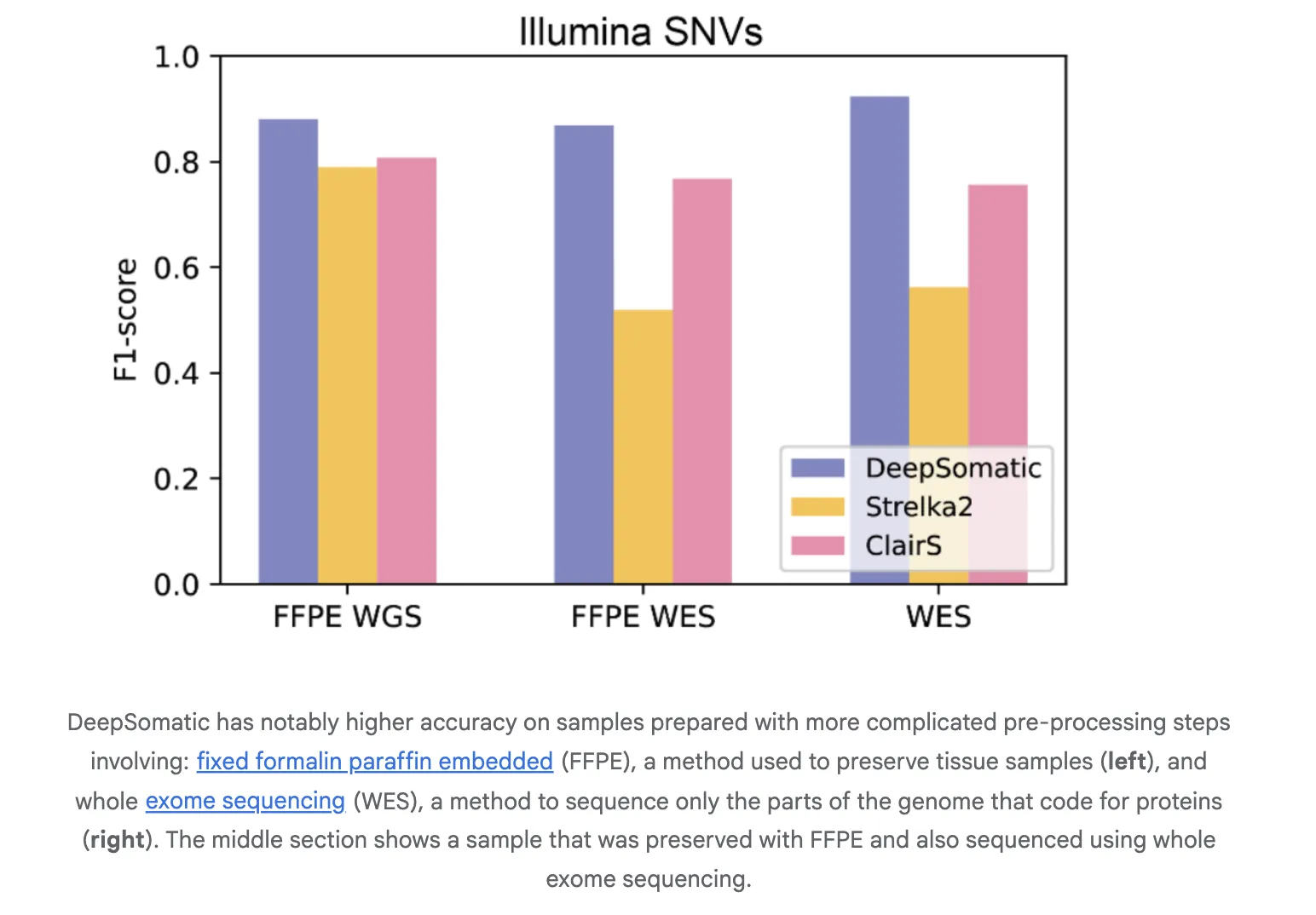

Generalize to real samples
The team evaluated cancer metastasis outside of the training set. Glioblastoma samples show known drivers restored. Childhood leukemia samples were tested only for tumor patterns and could not obtain clean normal values. The tool recovers known calls and reports other variants in the queue. These studies demonstrate that the representation and training scheme generalize to novel disease contexts and environments without matching normal values.
Main points
- DeepSomatic detects somatic SNVs (single nucleotide variants) and indels in Illumina, PacBio HiFi and Oxford Nanopore and is based on the DeepVariant method.
- The pipeline supports tumor-normal and tumor-only workflows, includes FFPE WGS and WES models, and is published on GitHub.
- It encodes the stack of reads into a tensor-like image and uses a convolutional neural network to classify somatic sites and emit VCF or gVCF.
- Training and evaluation were performed using the CASTLE dataset, which contains 6 matched tumor-normal cell line pairs sequenced on the three platforms and provides baselines and joins.
- The reported results show that the F1 indel rate is approximately 90% on Illumina and over 80% on PacBio, which is better than common baselines, with 329,011 individual cell variants identified in the reference sample.
DeepSomatic is a practical step toward somatic variant calling across sequencing platforms. The model retains DeepVariant’s image tensor representation and convolutional neural networks, so the same architecture can be extended from Illumina to PacBio HiFi to Oxford Nanopore with consistent preprocessing and output. The CASTLE dataset is the right move, providing matched tumor and normal cell lines via 3 techniques, thereby enhancing training and benchmarking and aiding reproducibility. The reported results highlight indel accuracy, with an F1 of approximately 90% on Illumina and over 80% on PacBio relative to lower baselines, which addresses a long-standing weakness in indel detection. The pipeline supports WGS and WES, normal tumors and tumors only, and FFPE, consistent with practical laboratory constraints.
Check Technical papers, technical details, datasets and GitHub repository. Please feel free to check out our GitHub page for tutorials, code, and notebooks. In addition, welcome to follow us twitter And don’t forget to join our 100k+ ML SubReddit and subscribe our newsletter. wait! Are you using Telegram? Now you can also join us via telegram.

Michal Sutter is a data science professional with a master’s degree in data science from the University of Padua. With a solid foundation in statistical analysis, machine learning, and data engineering, Michal excels at transforming complex data sets into actionable insights.
🙌 FOLLOW MARKTECHPOST: Add us as your go-to source on Google.


