BIOMNI-R0: New Agesic LLM end-to-end training with multiple turns augmented learning for biomedical research experts intelligence
AI’s role in biomedical research continues to grow
field Biomedical Artificial Intelligence Rapidly evolving, the demand for agents capable of performing leapfrog tasks is growing Genomics, clinical diagnosis and molecular biology. These agents are not only designed to retrieve facts. They’re hopeful Reasons for Complex Biological Problemsinterpret patient data and extract meaningful insights from a wide range of biomedical databases. Unlike general AI models, biomedical agents must interface with domain-specific tools, understand biological hierarchies, and simulate workflows similar to researchers to effectively support modern biomedical research.
Core Challenge: Matching Expert-level Inference
However, Achieve expert performance In these tasks, it is far from trivial. Most large language models lack when dealing with the nuances and depth of biomedical reasoning. They may succeed on surface-level retrieval or pattern recognition tasks, but often fail when challenged Multi-step reasoning,,,,, Diagnosis of rare diseasesor Gene prioritynot only the field of data access, but also the contextual understanding and the judgment of specific fields. This limitation creates a clear gap: How to train biomedical AI agents that can think and behave like fields like experts.
Why are traditional methods insufficient
Some solutions utilize Supervised learning In curated biomedical dataset or Search ceremony generation These methods have disadvantages for ground responses in literature or databases. They often rely on Static prompts and predefined behaviors that lack adaptability. Furthermore, many of these agents are working to effectively execute external tools and their The reasoning chain crashes When facing unfamiliar biomedical structures. This vulnerability makes them unsuitable Dynamic or high-risk environmentsinterpretability and accuracy in places that are not commercially acceptable.
Biomni-R0: New paradigm for using reinforcement learning
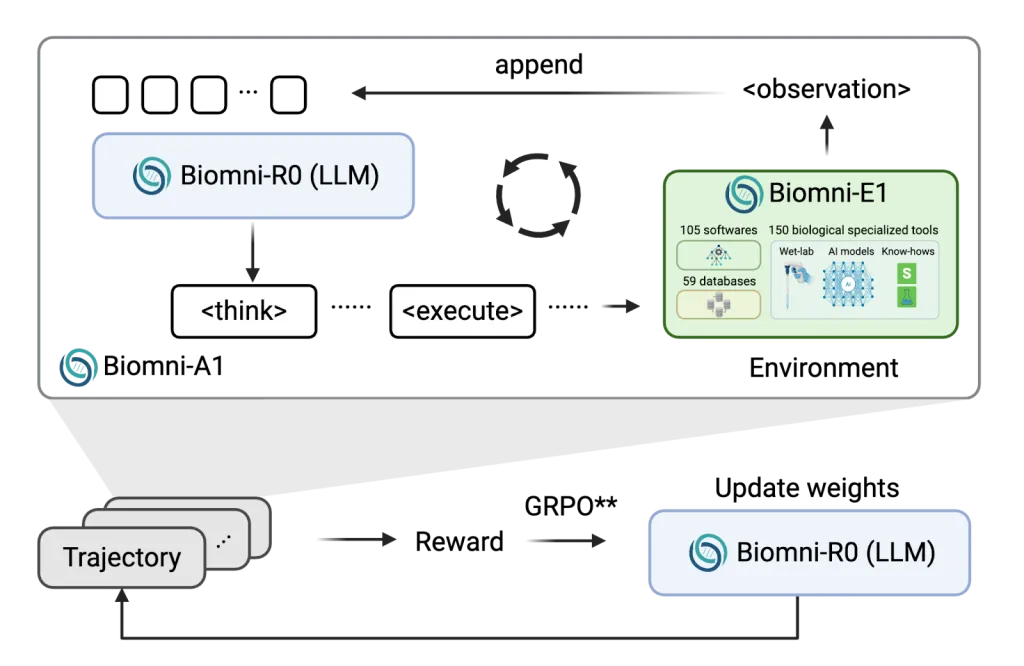
Researchers at Stanford University and UC Berkeley A new family of models is introduced called Biomni-R0through application Strengthening Learning (RL) to the Biomedical Agent Foundation. These models, Biomni-R0-8b and Biomni-R0-32baccepted RL environment tailored for biomedical reasoningusing expert logout tasks and novel reward structures. The collaboration combines Stanford University’s Biomni Agent and Environment Platform With the University of California, Berkeley Skyrl Reinforcement Learning Infrastructurea capability designed to push biomedical drugs to the human level.
Training strategies and system design
Research has introduced Two-stage training process. First, they used Supervised fine-tuning (SFT) The ability of the agent to follow a structured reasoning format is effectively guided on the high-quality trajectory of sampling from Claude-4 sonnets. Next, they use Reinforcement learningoptimize two rewards: one Correctness (e.g., select the correct gene or diagnosis), another Response format (For example, use structured
To ensure computing efficiency, the team developed Asynchronous launch plan Minimize bottlenecks caused by external tool delays. They also expanded Context length is 64K tokenallowing agents to effectively manage multi-step reasoning conversations.
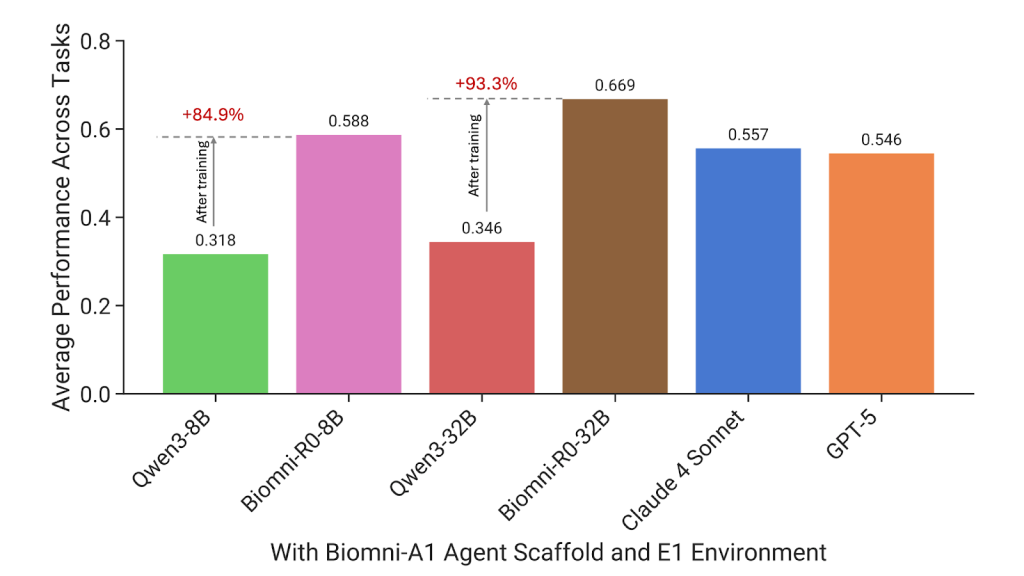
Results performed better than boundary model
Performance growth is huge. Biomni-R0-32b scores 0.669jumps from the base model 0.346. even Biomni-R0-8bsmaller version, score 0.588more than general models like this Claude 4 sonnets and GPT-5they are all much larger. According to the task, BIOMNI-R0-32B is 7 out of 10 taskswhile GPT-5 leads in 2, while Claude 4 is only in 1. One of the most compelling results is Diagnosis of rare diseasesWhere Biomni-R0-32b arrives 0.67compared with QWEN-32B 0.03, More than 20 times improvement. Similarly, in GWAS variant prioritythe model’s scores are from 0.16 arrive 0.74prove the value of domain-specific reasoning.

Design scalability and accuracy
Training large biomedical agents requires handling resource-heavy promotions involving external tool execution, database queries, and code evaluation. To manage this, the system breaks down Environment execution from Model inferenceallows for more flexibility to scale and reduce idle GPU time. This innovation ensures Effective use of resourceseven using tools with different execution delays. Longer inference sequences have also proven to be beneficial. RL-trained models are always produced Longer structured responsewhich is closely related to better performance, emphasizes The depth and structure of reasoning It is a key indicator for biomedical experts to understand.
Key points of research include:
- Biomedical agents must perform deep reasoningnot only searched in genomics, diagnostics and molecular biology.
- this Central issue Expert-level task performance is being achieved, mainly in complex areas such as rare diseases and genetic priorities.
- Traditional methodincluding supervised fine-tuning and search-based models, are often lacking in robustness and adaptability.
- Biomni-R0developed by Stanford University and the University of California, Berkeley Reinforcement learning With expert-based rewards and structured output formats.
- this Two-stage training pipeline,SFT is followed by RL, which is very effective in optimizing performance and inference quality.
- Biomni-R0-8b Provides strong results through smaller architectures, Biomni-R0-32b In 7 of 10 tasks, a new benchmark was set, better than Claude 4 and GPT-5.
- Reinforcement learning enables agents Generate longer, more coherent inference trajectoriesa key characteristic of expert behavior.
- This work is Super Expert Biomedical Agentable to accurately automate complex research workflows.
Check Technical details. Check out ours anytime Tutorials, codes and notebooks for github pages. Also, please stay tuned for us twitter And don’t forget to join us 100K+ ml reddit And subscribe Our newsletter.

Michal Sutter is a data science professional with a master’s degree in data science from the University of Padua. With a solid foundation in statistical analysis, machine learning, and data engineering, Michal excels in transforming complex datasets into actionable insights.


